As a postdoctoral investigate affiliate in the lab of BTI college member Frank Schroeder, Max Helf observed his labmates continually battle when they have been examining info. So, he resolved to do something about it and designed a free of charge, open-source app named Metaboseek, which is now essential to the lab’s perform.
The Schroeder lab studies the roundworm Caenorhabditis elegans, one particular of the most effective design programs for human biology, to learn new metabolites that govern evolutionarily conserved signaling pathways and could be useful as qualified prospects for the growth of new pharmaceuticals or agrochemicals. The scientists carry out this job by evaluating the metabolites among two different worm populations—a process named comparative metabolomics.
Given that samples routinely have far more than 100,000 compounds in them, computational techniques are critical to conduct the evaluation.
The workforce experienced been relying on software package deals that did not offer the essential stage of adaptability to very easily personalize investigation parameters. That limitation, and the deficiency of a suited graphical consumer interface, meant Helf’s colleagues confronted the cumbersome job of visually inspecting mounds of data—for example, to place doable fake positives—and jumping among several other software program equipment to affirm and filter out individuals meaningless final results.
“It just seemed really inefficient to me, and I could not get more than the shortcomings of other program solutions for this challenge,” Helf reported. “I thought there had to be an less complicated way, so I started out to publish code for my possess software program.”
Helf formulated the first model of his software in 2017, and continued to make improvements to it over the up coming two many years. “Besides addressing the troubles my labmates ended up by now facing, I talked to them about what else held them back—what they wished to do but were not even trying—and crafted all those capabilities in the application,” stated Helf, who is now a bioinformatics item manager at proteomics corporation Biognosys AG. “I wanted this new resource to be person-friendly and accessible to any one who does chemical biology.”
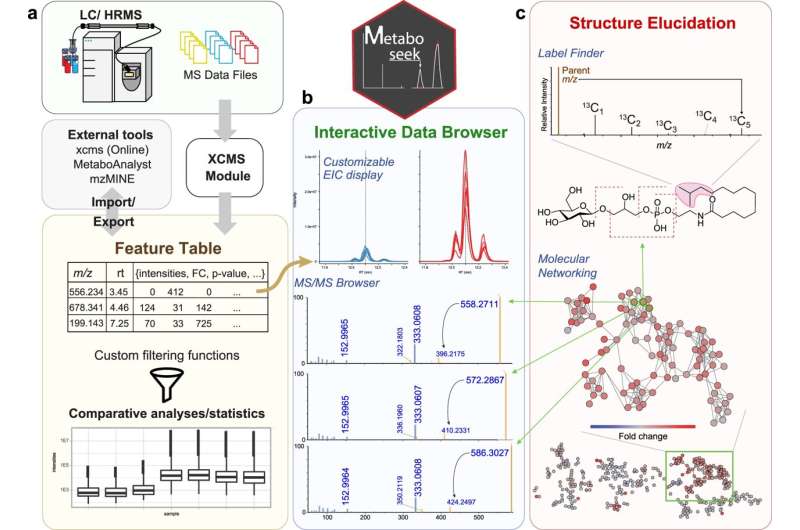
The final result was Metaboseek, an application with a graphical interface that incorporates several knowledge analysis tools that non-coding scientists would or else not have. The application streamlines the assessment of comparative metabolomics information by helping the researcher ascertain which info options are authentic and letting them dig deeper into those features—all within just the exact tool.
“Max did this without me even requesting it,” Schroeder claimed. “Just before I understood that this was occurring, there was Metaboseek. We began utilizing it, and now our lab and many collaborators couldn’t exist without it.”
In a study printed in Nature Communications on February 10, Schroeder’s crew provided evidence-of-notion for Metaboseek by implementing it to an vital fat fat burning capacity pathway that hadn’t nonetheless been studied: the α-oxidation pathway in C. elegans that helps crack down a course of fatty acids.
Making use of Metaboseek, the crew discovered that roundworms missing a critical gene in the α-oxidation pathway gathered hundreds of previously unreported metabolites. The findings are significant for the reason that α-oxidation is a essential biochemical pathway in worms that is conserved in individuals, Schroeder claimed.
“Bennett Fox did the chemistry work, so this examine was a wonderful collaboration amongst the two postdocs,” included Schroeder, who is also professor in Cornell University’s Division of Chemistry and Chemical Biology.
In accordance to Schroeder and Helf, there are a number of good reasons why there are not a large amount of fantastic analytic equipment for evaluating metabolomics info. Initial, comparative metabolomics is a comparatively young field as opposed with other information-significant fields of biology like genomics (which focuses on DNA) and proteomics (which focuses on proteins), so there has not been sufficient time to establish application instruments and databases infrastructure.
In addition, about the last ten years, the advent of affordable, ultra-large-resolution mass spectrometers for amassing metabolomics info has elevated by probably much more than tenfold the quantity of knowledge one sample can generate—creating an even greater need for advanced resources that can maintain up with the flood of info.
Metaboseek fulfills these desires with an array of features for examining a variety of sorts of details to support compound identification, construction determination, assignment of metabolites to family members based on structural similarities, tracking radiolabeled compounds, and a lot more.
An R offer for detailed info examination of peptide- and protein-centric bottom-up proteomics details
Maximilian J. Helf et al, Comparative metabolomics with Metaboseek reveals features of a conserved unwanted fat metabolic process pathway in C. elegans, Character Communications (2022). DOI: 10.1038/s41467-022-28391-9
Citation:
New software package to aid find out useful compounds (2022, March 29)
retrieved 30 March 2022
from https://phys.org/information/2022-03-application-worthwhile-compounds.html
This document is subject to copyright. Aside from any honest working for the function of non-public examine or study, no
section might be reproduced without having the prepared authorization. The articles is offered for facts needs only.
